I demonstrated that the vast majority (>93%) of plasmids in the Addgene repository contain fragments of ORFs. These are often likely just “dead weight”, taking up space that could be used in other parts of the plasmid. However, at least in some plasmids, these protein fragments appear to downstream of intact constitutive promoters. The figure below shows one of the most commonly requested plasmids from Addgene; on the left is the annotation that Addgene provides (via Snapgene), and on the right is my annotation.
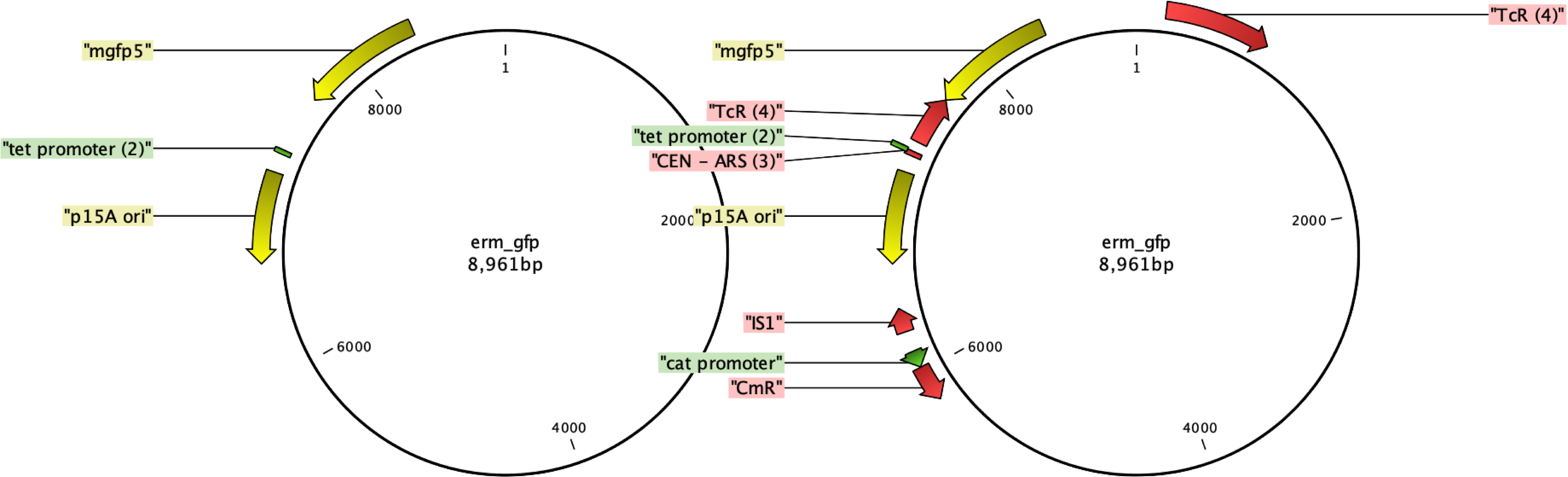
While most of the major components are identified in the Addgene annotation (origin, promoter, GFP), a chloramphenicol resistance gene fragment is missed. Since this fragment is downstream of a constitutive promoter, this fragment should be constitutive expressed, where the fragment is a chimera of part chloramphenicol efflux pump, and part random peptide gibberish. This could be quite toxic to the cell. My annotation also shows discarded fragments of tetracycline resistance. Strikingly, either of the annotation methods leaves a large portion of the plasmid unannotated. This is because this plasmid encodes for erythromycin resistance genes, which are not commonly found in engineered plasmids. This highlights improvements that can be made on my annotation pipeline.

